Research
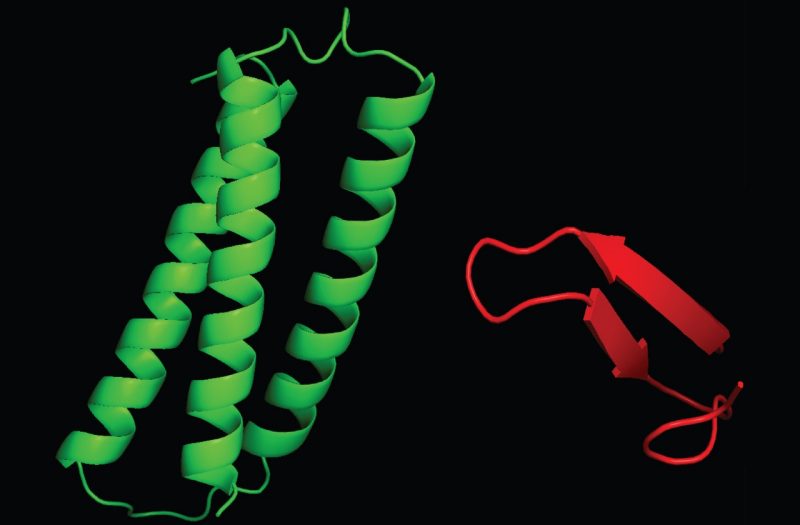
Our research goal is to understand how protein domains transduce signals from biological membranes. Our laboratory employs biophysical approaches - including high field nuclear magnetic resonance spectroscopy, isothermal calorimetry, circular dichroism, computer modeling, liposome-binding assays, fluorescence spectroscopy and surface plasmon resonance spectroscopy - to determine protein:lipid interfaces, ligand binding pockets and membrane insertion of protein domains from the molecular to the atomic resolution. We validate our functional and structural approaches by using normal and disease-associated cell lines. With these tools we can establish how protein-ligand interactions control the function of proteins by modulating their subcellular localization.
Role of Disabled-2 in Sulfatide-mediated Platelet Aggregation
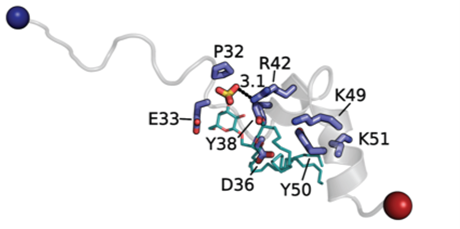
Disabled-2 (Dab2) is an adaptor protein that regulates platelet aggregation through two distinct mechanisms. In the first, Dab2 acts intracellularly to downregulate the integrin αIIbβ3 receptor, shifting it to a low-affinity state and thereby reducing adhesion and aggregation. In the second mechanism, Dab2 is released extracellularly, where it interacts with key pro-aggregatory mediators—specifically, the integrin αIIbβ3 receptor and sulfatides-to block their association with fibrinogen and P-selectin, respectively.
Our previous research identified a 35-amino-acid region within Dab2, referred to as the sulfatide-binding peptide (SBP), which contains two potential sulfatide-binding motifs characterized by consecutive polybasic regions. Using molecular docking, nuclear magnetic resonance, lipid-binding assays, and surface plasmon resonance, we determined the key Dab2 residues within SBP that mediate sulfatide binding. Molecular docking analyses revealed that a hydrophilic region centered around R42 interacts with the sulfatide headgroup, whereas the C-terminal polybasic region contributes to interactions with the lipid acyl chains. Moreover, we demonstrated that R42 in Dab2 SBP plays a major role in inhibiting platelet P-selectin surface expression.
Interestingly, the sulfatide-interacting residues within Dab2 SBP resemble those found in other sphingolipid-binding proteins, suggesting a conserved mechanism of lipid recognition among sulfatide-binding proteins. This project is being carried out in collaboration with Prof. Carla Finkielstein and Anne Brown (Virginia Tech).
Role of Modular Proteins in Endosomal Protein Trafficing
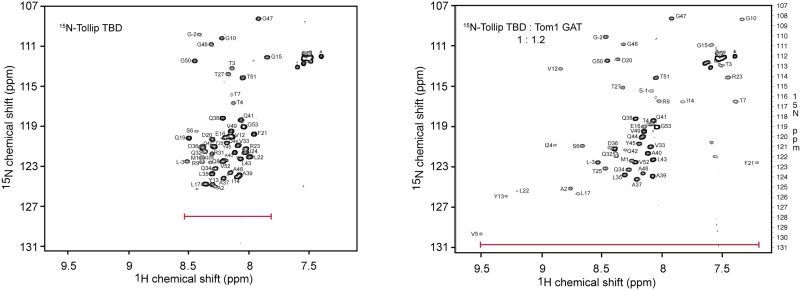
Ubiquitylation is a tightly regulated post-translational modification in which proteins are conjugated with either monoubiquitin or polyubiquitin chains. These ubiquitin modifications are recognized by ubiquitin-binding domains present in various effector proteins. Our research focuses on the structural characterization of two adaptor proteins, TOLLIP and TOM1, which assemble into a complex known as the alternative ESCRT-0 complex, responsible for transporting ubiquitylated cargo within endosomal compartments.
We have previously shown that the central C2 domain of TOLLIP preferentially interacts with phosphoinositides, a key interaction required for its membrane localization. Intriguingly, we discovered that ubiquitin downregulates TOLLIP’s lipid-binding. Specifically, we observed an ubiquitin dose-dependent inhibition of TOLLIP’s association with phosphoinositides, mediated by blocking the interaction between the TOLLIP C2 domain and phosphoinositide lipids. These findings led us to hypothesize that the TOLLIP C2 domain functions as a novel ubiquitin-binding domain. Also, we have obtained detailed structural insights into the formation of the TOM1-TOLLIP complex. Their interaction is mediated by the TBD and the C2 domains of TOLLIP, and the GAT domain of TOM1.
In addition, we identified an adjacent DXXLL motif-containing region to the TOM1 VHS domain, which enhances its affinity for ubiquitin and can be modulated by phosphorylation. TOM1 is an endosomal phosphatidylinositol 5-phosphate (PtdIns5P) effector under Shigella flexneri infection. We pinpointed a consensus PtdIns5P-binding motif in the VHS domain. We show that PtdIns5P binding by TOM1 is pH-dependent, similarly observed in its binding partner TOLLIP. Under acidic conditions, TOM1 retained its complex formation with TOLLIP, but was unable to bind ubiquitin. S. flexneri infection inhibits pH-dependent endosomal maturation, leading to reduced protein degradation.
We propose a model wherein pumping of H+ to the cytosolic side of endosomes contributes to the accumulation of TOM1, and possibly TOLLIP, at these sites, thereby promoting PtdIns5P- and pH-dependent signaling, facilitating bacterial survival. These projects are in collaboration with Carla Finkielstein (Biological Sciences, Virginia Tech), Anne Brown (Biochemistry, Virginia Tech), Samppa Ryhänen, Elina Ikonen, and Heljä Lång (New Children Hospital, University of Helsinki, Finland).
Role of Adaptor Proteins in Macropinocytosis
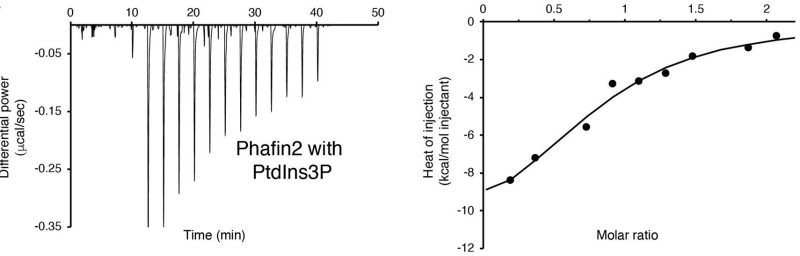
Macropinocytosis is a cellular process that enables the nonselective uptake of fluids and macromolecules, playing a critical role in cellular homeostasis, immune responses, pathogen entry, and disease progression. Phosphoinositides are central to this process, recruiting adaptor proteins required for macropinosome formation and maturation. One such adaptor, Phafin2, has emerged as a key regulator of macropinocytosis. Phafin2 exhibits two distinct phases of membrane association, suggesting it integrates phosphoinositide signaling with protein interactions to finely control macropinosome dynamics.
However, the precise mechanisms by which Phafin2 binds membranes and interacts with other macropinosomal proteins remain poorly understood. Phafin2 contains a PH domain, which binds phosphatidylinositol 3-phosphate (PtdIns3P) and phosphatidylinositol 4-phosphate (PtdIns4P), and a FYVE domain, which specifically recognizes PtdIns3P. Our previous work indicates that the FYVE domain constitutively binds PtdIns3P, while the PH domain is autoinhibited by the C-terminal poly-aspartic (polyD) motif. We propose that the FYVE domain modulates PH domain activity during macropinocytosis and that phosphoinositides orchestrate Phafin2’s interactions with macropinosomal proteins throughout this process. Understanding these mechanisms will provide insights into how cells regulate macropinocytosis and its implications in health and disease.

Daniel G. S. Capelluto, PhD
Professor, Biological Sciences Dept.
Fralin Life Sciences Institute
Office: 1015 Life Science Circle, Room 263C
Lab rooms: 250/252/254
Blacksburg, VA 24061-0477
capellut@vt.edu
(540) 231-0974



